Week 11: Categorical Predictors & Logistic Regression
Date: November 3, 2025
Today…
Regression
3 main reasons for using regression:
- As a description (what is the average salary for men and women?)
- As part of causal inference (Does being a woman result in a lower salary?)
- For prediction (“What happens if…” questions)
Explaining Variance

Model Interpretation
Once we have a model, we will be able to interpret the coefficients. For a bivariate regression, this was fairly straightforward
We would look to the beta (effect size) and interpret it as: for every 1 unit increase in our X variable, there will be beta units increase in our Y variable.
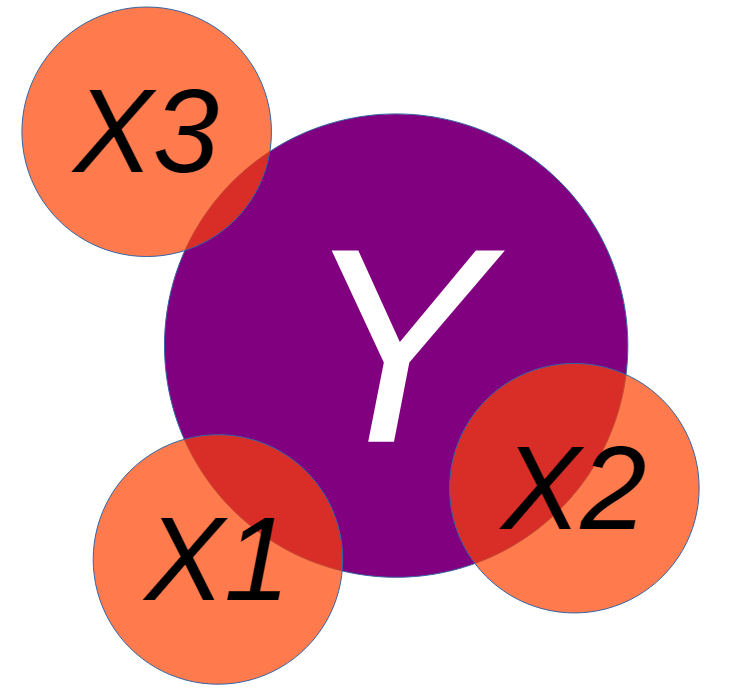
Multiple Regression
We may want to include additional predictors into our model to best explain the variance in an outcome:
\[ Y_i = b_0 + b_1X_{1i} + b_2X_{2i} + ... + b_nX_{ni}+ e_i \]
Explaining Variance
Model Interpretation
Since we have multiple predictors in the model, we have to interpret each predictor estimate while considering the other variables.
We would look to the beta (effect size) and interpret it as: for every 1 unit increase in our \(X_1\) variable, there will be beta units increase in our Y (outcome) variable, holding all other variables constant.
Reporting Standardized or Unstandardized?
INTERPRET b:
When the variables are measured in a meaningful metric
To compare the relative effects of different predictors in the same sample
INTERPRET β:
When the variables are not measured in a meaningful metric
To compare effects across samples or studies
Multiple Regression: Example
Previously, we predicted the # of Transformers movies someone watched by their age, income and # of books read
The regression equation would look something like this:
\[ Y = b_0 + b_1X_1 + b_2X_2 + b_3X_3 + e \]
Transformers Regression: \(b\)
Interpret:
Call:
lm(formula = transformers ~ age + income + books, data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-2.0283 -1.0658 -0.6646 0.8359 4.2314
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.6872892571 0.3197311424 8.405 0.000000000000000644 ***
age -0.0256880349 0.0054919383 -4.677 0.000003904370103857 ***
income -0.0000021955 0.0000009953 -2.206 0.0279 *
books 0.0018608303 0.0009792731 1.900 0.0581 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.504 on 427 degrees of freedom
(569 observations deleted due to missingness)
Multiple R-squared: 0.06449, Adjusted R-squared: 0.05791
F-statistic: 9.811 on 3 and 427 DF, p-value: 0.000002857Transformers Regression: \(\beta\)
scale()
Call:
lm(formula = scale(transformers) ~ scale(age) + scale(income) +
scale(books), data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-1.3007 -0.6834 -0.4262 0.5361 2.7135
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.04536 0.04817 -0.942 0.3469
scale(age) -0.27392 0.05856 -4.677 0.0000039 ***
scale(income) -0.10176 0.04613 -2.206 0.0279 *
scale(books) 0.09053 0.04764 1.900 0.0581 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.9642 on 427 degrees of freedom
(569 observations deleted due to missingness)
Multiple R-squared: 0.06449, Adjusted R-squared: 0.05791
F-statistic: 9.811 on 3 and 427 DF, p-value: 0.000002857Transformers Regression: \(\beta\)
lm.beta()
Call:
lm(formula = transformers ~ age + income + books, data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-2.0283 -1.0658 -0.6646 0.8359 4.2314
Coefficients:
Estimate Standardized Std. Error t value
(Intercept) 2.6872892571 NA 0.3197311424 8.405
age -0.0256880349 -0.2194418839 0.0054919383 -4.677
income -0.0000021955 -0.1036558341 0.0000009953 -2.206
books 0.0018608303 0.0890938373 0.0009792731 1.900
Pr(>|t|)
(Intercept) 0.000000000000000644 ***
age 0.000003904370103857 ***
income 0.0279 *
books 0.0581 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.504 on 427 degrees of freedom
(569 observations deleted due to missingness)
Multiple R-squared: 0.06449, Adjusted R-squared: 0.05791
F-statistic: 9.811 on 3 and 427 DF, p-value: 0.000002857How would we write up the results?
Call:
lm(formula = transformers ~ age + income + books, data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-2.0283 -1.0658 -0.6646 0.8359 4.2314
Coefficients:
Estimate Standardized Std. Error t value
(Intercept) 2.6872892571 NA 0.3197311424 8.405
age -0.0256880349 -0.2194418839 0.0054919383 -4.677
income -0.0000021955 -0.1036558341 0.0000009953 -2.206
books 0.0018608303 0.0890938373 0.0009792731 1.900
Pr(>|t|)
(Intercept) 0.000000000000000644 ***
age 0.000003904370103857 ***
income 0.0279 *
books 0.0581 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.504 on 427 degrees of freedom
(569 observations deleted due to missingness)
Multiple R-squared: 0.06449, Adjusted R-squared: 0.05791
F-statistic: 9.811 on 3 and 427 DF, p-value: 0.000002857How would we write up the results?
Code
| # of Transformers Movies | |||||
|---|---|---|---|---|---|
| Predictors | Estimates | std. Beta | CI | standardized CI | p |
| Intercept | 2.69 | 0.00 | 2.06 – 3.32 | -0.09 – 0.09 | <0.001 |
| Age | -0.03 | -0.22 | -0.04 – -0.01 | -0.31 – -0.13 | <0.001 |
| Income | -0.00 | -0.10 | -0.00 – -0.00 | -0.20 – -0.01 | 0.028 |
| # of Books | 0.00 | 0.09 | -0.00 – 0.00 | -0.00 – 0.18 | 0.058 |
| Observations | 431 | ||||
| R2 / R2 adjusted | 0.064 / 0.058 | ||||
A linear regression model was fit to the data to predict # of Transformers movies watched with participant age, income and reported # of books read. The model explains a significant, but small portion of variance in the outcome (F(3, 427) = 9.81, p < .001, adj. \(R^2\) = .06). Participant age ( \(\beta\) = -0.22, p < .001) and income ( \(\beta\) = -0.10, p = .028) were significantly related to the number of transformers movies, while the number of books was not (p = .508). These findings suggest that as individuals age, they tend to watch fewer transformers movies, controlling for income and # of books. Additionally, as income increases, we see a decrease in # of transformer movies while taking into account participant age and # of books.
But what about categorical variables as predictors?

Categorical Predictors
Typically identified by a grouping variable that may be a character
chr [1:1000] "Democrat" "Democrat" "Independent" "Republican" "Democrat" ...Need to change the variable from character to factor which will assign a number to each group
Categorical Data
Nominal - the numbers have no relation to the category it represents (Biological Sex)
- Biological Sex - Male (1), Female (2), Intersex (3)
Ordinal - the order of the numbers matters
- Education Levels - High school (1), Bachelor’s (2), Master’s (3), Doctorate (4)
Categorical Data - Regression
Can we use the numeric values that each category represents in a regression? I mean we can do just about anything. But, we should not use the numeric values as they are
Why can’t we do this?
Tip
Consider how we interpret the beta coefficients for slope
Categorical Data - Regression
Regressions assume that there are equal spacing between each of the values and that the spacing is meaningful
Going from High School to Bachelor’s is considered equal to Master’s to Doctorate?
What does it mean to interpret Male to Female to Intersex?
To properly investigate the differences between the groups, we have to “trick” our regression equation - Enter Dummy Variables/Coding
Dummy Coding
Numerical placeholders used to represent categorical variables
Taking a categorical variable with \(k\) levels (e.g., Democratic, Independent, Republican) into \(k-1\) binary variables.
| political_affiliation | becomes | Ind (binary1) | Rep (binary2) |
|---|---|---|---|
| Democrat | –> | 0 | 0 |
| Independent | –> | 1 | 0 |
| Republican | –> | 0 | 1 |
| … | … | … |
Dummy Coding
For the variable with \(k\) levels, we use \(k-1\) binary variables…why not use all of them?
Including all three variables would result in perfect multicollinearity. All Democrats would be related to other Democrats and unrelated to everything else
By including 2 binary variables, we are able to obtain all information about group membership
Dummy Coding
The group that has 0’s for all the binary variables is the reference group
For interpretation, we will make our statements in reference to this group
| political_affiliation | becomes | Ind (binary1) | Rep (binary2) |
|---|---|---|---|
| Democrat | –> | 0 | 0 |
| Independent | –> | 1 | 0 |
| Republican | –> | 0 | 1 |
| … | … | … |
Categorical Regression
Going back to our Transformers dataset, let’s see how our political affiliation variable can predict # of transformers movies
Usually you have to create the separate dummy variables, but not in R
You can also double check the dummy/contrast coding
Interpretation
We are shown the different levels of our predictor variable, but you will not see a predictor for the reference group
Tip
Think about how we interpret the intercept of a regression model. What are all values of our predictors set to be when looking at the intercept estimate?
Call:
lm(formula = transformers ~ pol, data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-1.3447 -1.1804 -0.3447 0.8196 3.8367
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.18039 0.09399 12.559 <0.0000000000000002 ***
polIndependent 0.16434 0.12349 1.331 0.184
polRepublican -0.01713 0.14257 -0.120 0.904
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.501 on 799 degrees of freedom
(198 observations deleted due to missingness)
Multiple R-squared: 0.003244, Adjusted R-squared: 0.0007487
F-statistic: 1.3 on 2 and 799 DF, p-value: 0.2731Refactor Reference Group
Maybe we want to compare to a specific group
We need to then update the reference group
Call:
lm(formula = transformers ~ pol2, data = cah_data)
Residuals:
Min 1Q Median 3Q Max
-1.3447 -1.1804 -0.3447 0.8196 3.8367
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.34473 0.08011 16.786 <0.0000000000000002 ***
pol2Democrat -0.16434 0.12349 -1.331 0.184
pol2Republican -0.18146 0.13383 -1.356 0.175
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.501 on 799 degrees of freedom
(198 observations deleted due to missingness)
Multiple R-squared: 0.003244, Adjusted R-squared: 0.0007487
F-statistic: 1.3 on 2 and 799 DF, p-value: 0.2731lm() or aov()
Didn’t this look similar to something we’ve done before?
We are examining group differences in Transformer Movies (continuous) by Political Affiliation (group/categorical)
IT IS JUST AN ANOVA
Everything is a linear model
We are able to come to the same conclusions that there are no significant differences between groups
Break ☕🍵🥐
Activity Time 🎨
Categories in Action
Goal: Gain greater familiarity with dummy coding and categorical predictors
Scenario: We are researchers examining the impact of a new intervention on reducing the vocalization “6️⃣7️⃣” in the youths. We have done classroom observations to collect data on how many times students say “6️⃣7️⃣” after receiving the intervention. This is the data you have in front of you. Youths have been randomly assigned to one of 3 groups, and we need to determine which had the biggest impact.
Note
You will submit one Rmd file for the whole group. Be sure to include all group member’s names.
Part 1
- Get into groups of 4
- Download the data [Download Categorical data (.csv)]
- Enter the data into the spreadsheet with the chips that you have
- Each chip has an ID number (three digits) and a score (between 5 - 20)
- These are the scores for the “post”
- Be sure to identify their Group (the color of the chip)
Part 2
Import the data into R
- You will only have data for one school (be sure to
filter()to include your data)
- You will only have data for one school (be sure to
On the board, put a portion of your data in a table along with your Dummy Code Scheme
- Dr. Haraden should have different sections designated, if not, please remind him nicely (he’s doing his best)
Part 3
Calculate the average 6️⃣7️⃣ Score for your reference group:
Average Score for Group 1 (Reference) = ______________ = \(\beta_0\)Calculate the average 6️⃣7️⃣ Score for your other two groups:
Average Score for Group 2 = ______________
Average Score for Group 3 = ______________Calculate the “Coefficients” by finding the mean difference from the reference group:
\(\beta_1\) = (Avg. Group 2 Score) - (Avg. Group 1 Score) = __________
\(\beta_2\) = (Avg. Group 3 Score) - (Avg. Group 1 Score) = __________
Part 4
Write out the regression equation for the model in which we are predicting 6️⃣7️⃣ Scores with our Dummy Variables. It should follow a format that we’ve used before ( \(Y = b_0 + b_1X_1 + b_2X_2 + e\) ), but replacing your estimates in the correct places with the appropriate variables.
Once your group agrees on the equation, write it on the board for your group
Categories in Action: Using R
Now that we have the data entered appropriately, let’s double check, and include some additional variables!
Import dataset, confirm predictions, include pre-scores to see how that changes interpretation
Warning
Dr. Haraden is going to start opening up R and doing a follow-along thing. You have been warned
Everything is a linear model
Maybe another break?
Introducing Generalized Linear Model
6️⃣7️⃣ Status
In our last example, we were able to see how these categorical variables could predict a continuous variable. This is perfect for linear regression.
What if we want to see if students have “recovered” from 6️⃣7️⃣?
We would then ask: ““Which participants dropped below the clinical threshold for 6️⃣7️⃣ at follow-up? Now, our outcome is either recovered or not recovered.”
Recovery Status
Now we have a binary outcome; Yes/No recovery
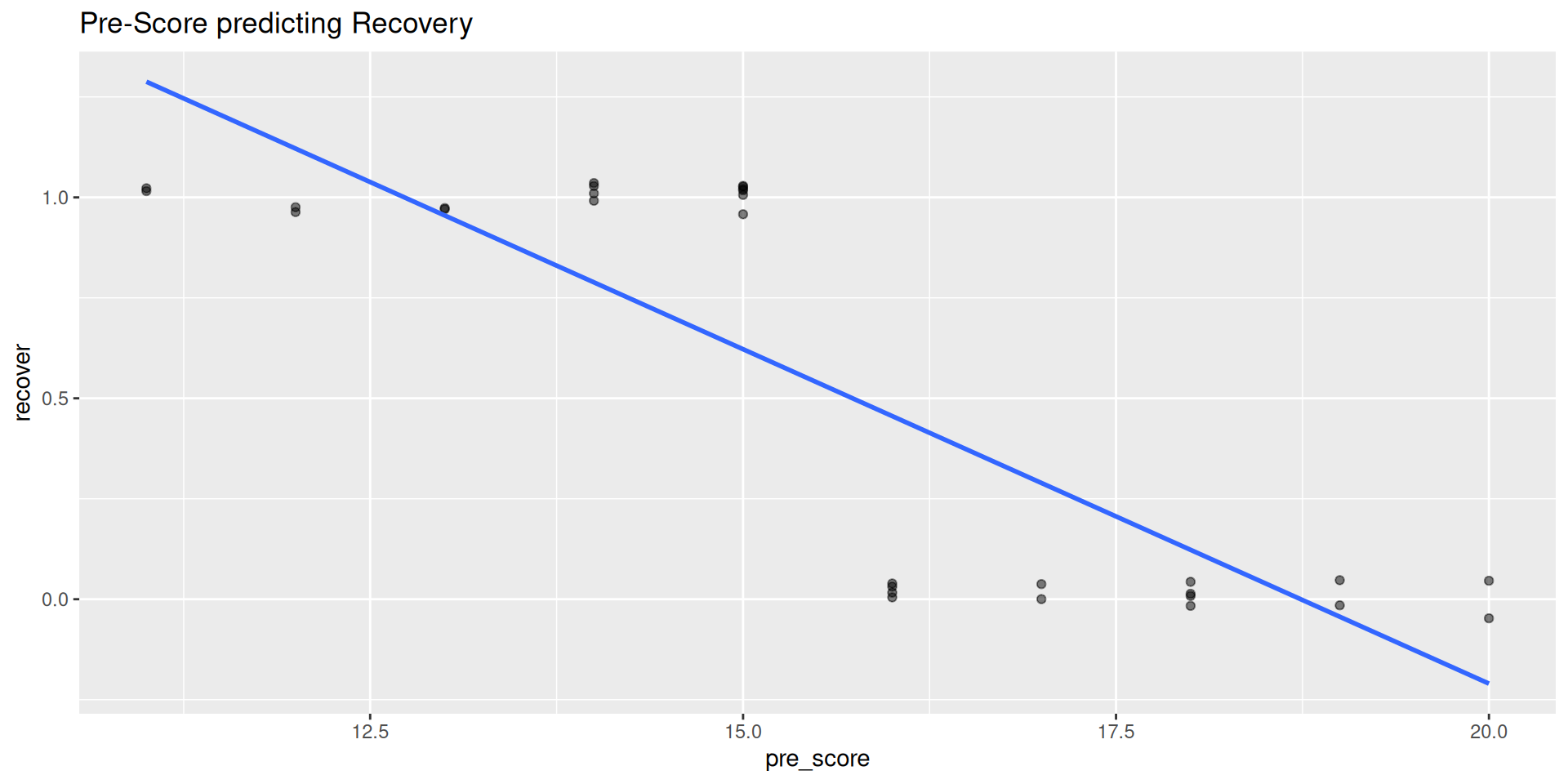
What happens when we fit a linear regression? What are the chances of someone with a pre-score of 12 recovering by the follow-up?
Code
cat_reg <- import(here("files", "data",
"cat_reg_complete.xlsx")) %>%
janitor::clean_names() %>%
# 1 = recovered; 0 = not recovered
mutate(recover = if_else(post_score > 12, 0, 1))
cat_reg %>%
ggplot(aes(pre_score, recover)) +
geom_jitter(width = 0, height = 0.05, alpha = 0.5) +
geom_smooth(method = "lm", se = FALSE) +
labs(
title = "Pre-Score predicting Recovery"
)
Probability to Odds to Log-Odds
Probability (p): The chance of an event happening. Ranges from 0 to 1
Odds: The ratio of the probability of an event happening to it not happening.
\(Odds = \frac{p}{1-p}\)
Ranges from 0 to ∞. An odds of 4 means the event is 4 times more likely to happen than not.
Log-Odds (logit): The natural log of the odds
\(Logit(p) = ln(\frac{p}{1-p})\)
Ranges from -∞ to +∞
Important
This step transforms our bounded outcome variable (0/1) to an unbound one!
Generalized Linear Model (GLM)
A generalization of a linear model (duh) that is used when the response variable has a non-normal error distribution
Most commonly used when there is a binary (0-1) or count variable as the outcome (we will focus on the binary)
Ultimately, we are trying to identify the probability of the outcome taking the value 1 (“success”) that is being modeled in relation to the predictor variables
GLM: Logistic Regression
\[ transformation(p_i)=\beta_0+\beta_1x_{1,i} + \beta_2x_{2,i} + \cdots+\beta_lx_{k,i} \]
We have to apply a transformation to the left side so that it can take variables beyond just 0 & 1
A common transformation is the \(logit\ transformation\)
\[ \log_{e}\left( \frac{p_i}{1-p_i} \right) = \beta_0 + \beta_1 x_{1,i} + \beta_2 x_{2,i} + \cdots + \beta_k x_{k,i} \]
GLM - CAH data 👻
Research Question: Does age, the number of books and the number of transformer movies predict the belief in ghosts?
GLM - CAH data Interpretation
Note
Interpretation is still the same as linear regression, except we are dealing with log-odds of the outcome. What does that mean??
Call:
glm(formula = ghosts ~ age + books + transformers, family = "binomial",
data = cah_data)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.5175834 0.2466775 -2.098 0.035886 *
age -0.0046259 0.0044171 -1.047 0.294975
books 0.0008966 0.0009700 0.924 0.355311
transformers 0.1643321 0.0453671 3.622 0.000292 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 1195.3 on 900 degrees of freedom
Residual deviance: 1177.0 on 897 degrees of freedom
(99 observations deleted due to missingness)
AIC: 1185
Number of Fisher Scoring iterations: 4GLM - CAH data Interpretation (tables)
Note
We want to be able to interpret these coefficients more easily so we put them into Odds Ratios
sjPlot is always coming in with the good tables
Odds Ratios (OR)
OR > 1: The predictor increases the odds of the outcome. (e.g., OR of 2.5 means the odds of believing in ghosts are 2.5 times higher).
OR < 1: The predictor decreases the odds of the outcome. (e.g., OR of 0.4 means the odds of believing in ghosts are 60% lower).
OR = 1: The predictor has no effect on the odds of the outcome.
Visualization
Extract the model implied probabilities for each individual
Plotting the predicted probabilities
probs %>%
ggplot(aes(age, .fitted)) +
geom_line(color = "blue", linewidth = 1.5) +
labs(
title = "Predicted Probability of Believing in Ghosts by Age",
subtitle = "Holding # of Books and Transformers Movies at their mean",
x = "Age (years)",
y = "Predicted Probability of Believing in Ghosts"
) +
ylim(0, 1) + # Keep the y-axis bounded at 0 and 1
theme_minimal()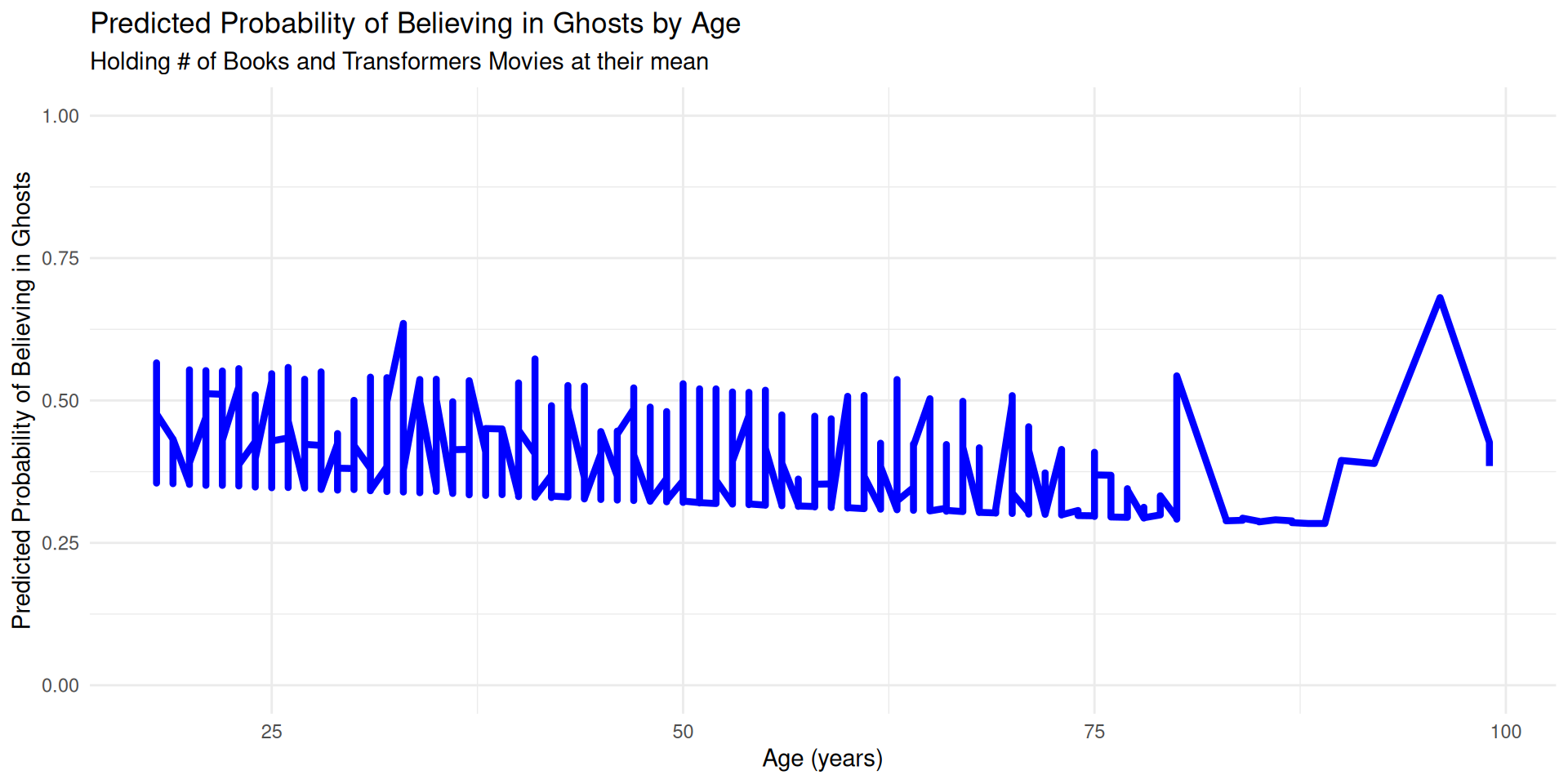
Logistic Regression: Summary
| Feature | Linear Regression | Logistic Regression |
|---|---|---|
| Outcome Variable | Continuous | Categorical (Binary) |
| Equation | \(Y=β_0+β_1X\) | \(ln(\frac{p}{1-p})=β_0+β_1X\) |
| Key Interpretation | \(\beta_1\) is the change in the mean of Y | \(\exp(\beta_1)\) is theodds ratio |
| R Function | lm() |
glm(..., family="binomial") |

