Week 03: Wrangling
Date: September 8, 2025
Today

Introduce: Library Tracker
There are so many things to remember. Let’s try to make it easier.
Use this page to document the different libraries that we are using in class and for the labs. Share any other libraries that you find a long the way!
Remember, this is for future you! Let’s try to be nice to them.
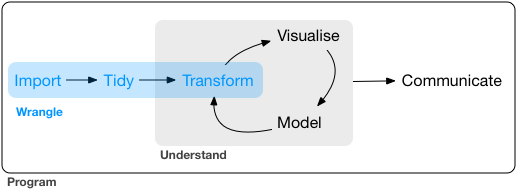
Starting Up
Let’s first start by opening our Project
Then, create a new Notebook/Markdown Document that we will use for today
Setup the libraries and bring in the data
- We will use the TIPI data from the last lab
[1] "ID" "Progress" "Duration (in seconds)"
[4] "Consent" "genderid" "genderid_7_TEXT"
[7] "sex" "age" "year_school"
[10] "Q85" "Q85_6_TEXT" "TIPI_1"
[13] "TIPI_2" "TIPI_3" "TIPI_4"
[16] "TIPI_5" "TIPI_6" "TIPI_7"
[19] "TIPI_8" "TIPI_9" "TIPI_10"
[22] "Sleep Quality" "Hours of Sleep" Naming Conventions
Remember in the lab when things were named all funky? We used the rename() function to do that last time. This can be helpful if you want to change it to something specific, but we may just want to make these names a little cleaner.
Introducing janitor https://sfirke.github.io/janitor/index.html
[1] "id" "progress" "duration_in_seconds"
[4] "consent" "genderid" "genderid_7_text"
[7] "sex" "age" "year_school"
[10] "q85" "q85_6_text" "tipi_1"
[13] "tipi_2" "tipi_3" "tipi_4"
[16] "tipi_5" "tipi_6" "tipi_7"
[19] "tipi_8" "tipi_9" "tipi_10"
[22] "sleep_quality" "hours_of_sleep" select()
.png)
Using select()
You used this in lab, so you are all experts. Let’s review by looking at the cheatsheet for dplyr.
The dplyr package makes data wrangling and transformation much easier. select() allows you to…well…select the columns that you want to keep.
Create a dataset that only includes: id, progress, duration, TIPI items
Using select()
filter()
.png)
Extract Rows
The ‘filter()’ function is used to subset observations based on their values.
The result of filtering is a data frame with the same number of columns as before but fewer rows.
The first argument is data and subsequent arguments are logical expressions that tell you which observations to retain in the data frame.
Note: You are stating which types of rows you want to keep. If a variable can answer TRUE to your condition, then it will stay in the data.
# A tibble: 38 × 14
name height mass hair_color skin_color eye_color birth_year sex gender
<chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 Darth V… 202 136 none white yellow 41.9 male mascu…
2 IG-88 200 140 none metal red 15 none mascu…
3 Bossk 190 113 none green red 53 male mascu…
4 Lobot 175 79 none light blue 37 male mascu…
5 Ackbar 180 83 none brown mot… orange 41 male mascu…
6 Nien Nu… 160 68 none grey black NA male mascu…
7 Nute Gu… 191 90 none mottled g… red NA male mascu…
8 Jar Jar… 196 66 none orange orange 52 male mascu…
9 Roos Ta… 224 82 none grey orange NA male mascu…
10 Rugor N… 206 NA none green orange NA male mascu…
# ℹ 28 more rows
# ℹ 5 more variables: homeworld <chr>, species <chr>, films <list>,
# vehicles <list>, starships <list>Filter Observations
We can now generate a subset of observations based on a particular value
There may be some data checks that you perform when wrangling data. One that I would suggest is to look at the overall completion percentages and the amount of time that it took for participants to complete the questionnaire.
This is exactly what filter() is set to do.
mutate()
.png)
Compute new variable
We often need to make a sum/mean score for a variable of interest, or transform it in some way.
The mutate() function is most commonly used to add new columns to your data frame that are functions of existing columns.
mutate() requires data as its first argument, followed by a set of expressions defining new columns.
- Note: New variables are automatically added at the end of the data frame (scroll to the right to see them)
For example, in the lab, we had the Ten Item Personality Inventory
Take a look at the scoring of the TIPI and compute the necessary variables for all subscales
Reverse Coding
There are some items in questionnaire data that are rated in the opposite direction from the other items. These need to be “reverse coded” or “reverse scored”
The TIPI documentions highlights certain variables that need this done
We can take the max value, add 1, then subtract the Item
tipi_2 tipi_2r tipi_4 tipi_4r
1 3 5 3 5
2 5 3 5 3
3 3 5 5 3
4 2 6 7 1
5 5 3 5 3
6 6 2 2 6Reverse Coding 2.0
With R, if there is something that is tedious, then that probably means that someone has made a library for it to make it easier.
Here comes reverse() from datawizard
datawizard::reverse()
We can try out this function with our TIPI data to see if we get something similar to before.
tipi_2 tipi_2r tipi_4 tipi_4r
1 3 5 3 5
2 5 3 5 3
3 3 5 5 3
4 2 6 7 1
5 5 3 5 3
6 6 2 2 6Sum Scores
And finally, we can use mutate() to create total scores (or really any type of computation)
For the TIPI data, we need to compute each of the 5 pieces of the Big Five (Extraversion, Agreeableness, Conscientiousness, Emotional Stability and Openness to Experience)
Exporting Data
export()
Now that we have a scored dataset, we will want to save that.
You can do this by re-running all of your steps above, or by exporting your dataset.
Note
Be careful when exporting and knitting docs. Each time you knit, it will run the export code. After I export, I usually will comment out that line of code.
