Intro to RMarkdown (and some regression stuff)
PSYC 640 - Fall 2023
Overview
Next couple of classes will be split between group work and R Markdown fancifulness
Presentations will start on Wednesday 12/6
Peer Review of Data analytic plan will be assigned tomorrow (how quickly can this be done?)
What are other things we can do? Or questions you have?
Last Time
- Multiple Regression
- Group work stuff
Today…
Review the regression stuff
RMarkdown!
- Even though we’ve been doing it this whole time
Data for Today
This data is taken from Kaggle and is simulated data to explore multiple linear regression. Variables include:
Scores from previous test
Average sleep hours
Average hours studied
Participating in Extracurriculars
# of Practice Questions used
Performance Index (Outcome)
Multiple Regression
Regression Equation
Going from this:
\[ \hat{Y} = b_0 + b_1X1 \]
To this
\[ \hat{Y} = b_0 + b_1X_1 + b_2X_2 + ... + b_kX_k \]
Regression coefficient values are now “partial” - Represents the contribution to all of outcome (\(\hat{Y}\)) from unique aspects of each \(X\)
Example - Single Predictor (tab_model())
Can customize the tables which we will focus more on in the next section
Call:
lm(formula = performance_index ~ hours_studied, data = student_perf)
Residuals:
Min 1Q Median 3Q Max
-33.667 -14.093 0.226 15.205 33.226
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 39.646 2.659 14.912 < 0.0000000000000002 ***
hours_studied 3.128 0.464 6.741 0.000000000157 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 17.67 on 205 degrees of freedom
Multiple R-squared: 0.1814, Adjusted R-squared: 0.1774
F-statistic: 45.43 on 1 and 205 DF, p-value: 0.0000000001569sjPlot::tab_model(fit1, show.std = TRUE,
title = "Figure 1",
pred.labels = c("Int.", "Hours Studied"),
dv.labels = "Performance Index")| Performance Index | |||||
| Predictors | Estimates | std. Beta | CI | standardized CI | p |
| Int. | 39.65 | -0.00 | 34.40 – 44.89 | -0.12 – 0.12 | <0.001 |
| Hours Studied | 3.13 | 0.43 | 2.21 – 4.04 | 0.30 – 0.55 | <0.001 |
| Observations | 207 | ||||
| R2 / R2 adjusted | 0.181 / 0.177 | ||||
Single Predictor - Visualizing
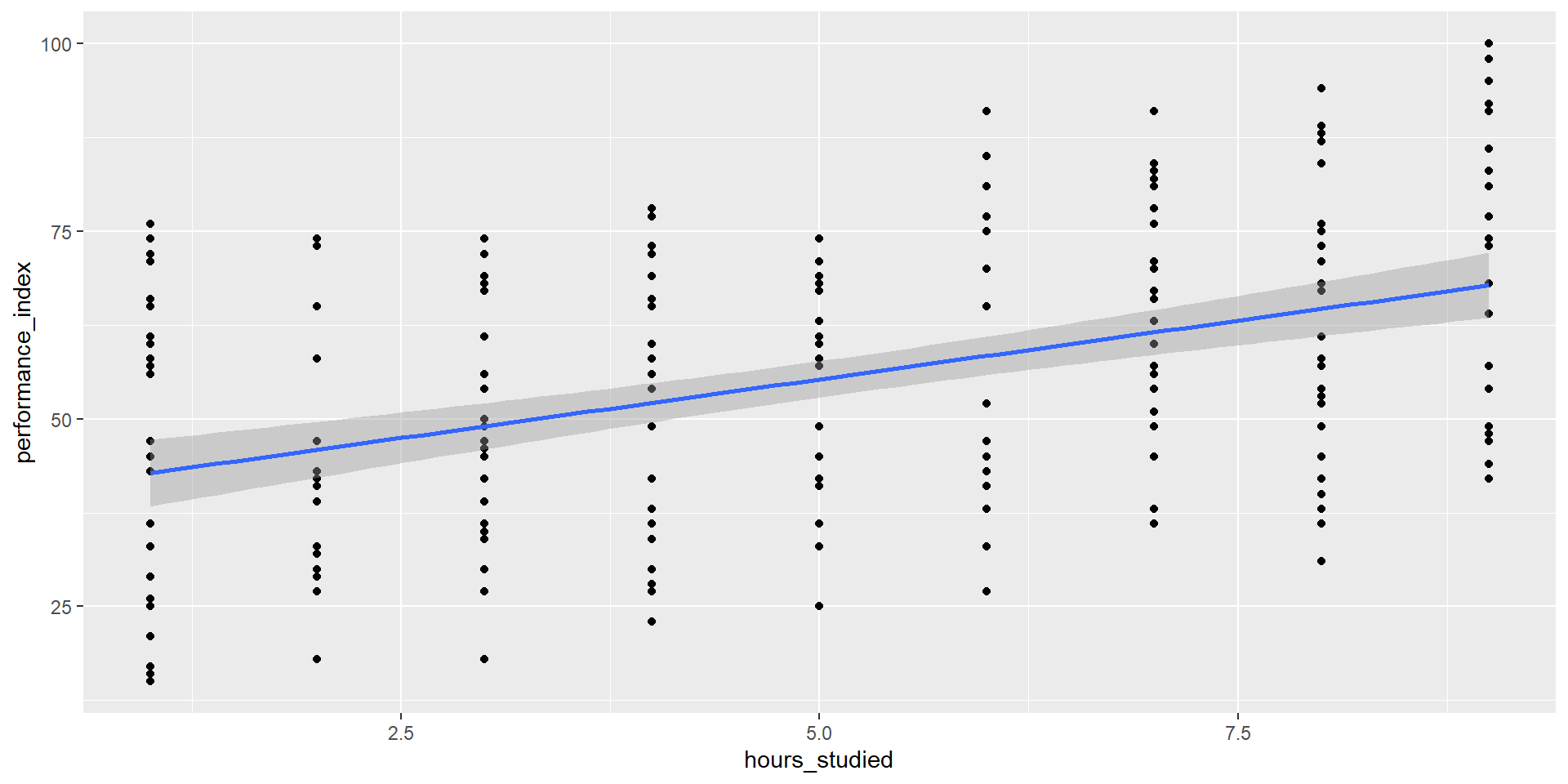
Single Predictor- Reporting
We fitted a linear model (estimated using OLS) to predict performance_index
with hours_studied (formula: performance_index ~ hours_studied). The model
explains a statistically significant and moderate proportion of variance (R2 =
0.18, F(1, 205) = 45.43, p < .001, adj. R2 = 0.18). The model's intercept,
corresponding to hours_studied = 0, is at 39.65 (95% CI [34.40, 44.89], t(205)
= 14.91, p < .001). Within this model:
- The effect of hours studied is statistically significant and positive (beta =
3.13, 95% CI [2.21, 4.04], t(205) = 6.74, p < .001; Std. beta = 0.43, 95% CI
[0.30, 0.55])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald t-distribution approximation.Multiple Predictors - Example
Now let’s add in some other variables that we think may contribute to a student’s “Performance Index”
fit2 <- lm(performance_index ~ hours_studied + sleep_hours + previous_scores,
data = student_perf)
summary(fit2)
Call:
lm(formula = performance_index ~ hours_studied + sleep_hours +
previous_scores, data = student_perf)
Residuals:
Min 1Q Median 3Q Max
-5.3665 -1.3541 -0.0376 1.3853 4.5946
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -33.159893 0.805496 -41.167 < 0.0000000000000002 ***
hours_studied 2.853131 0.052487 54.359 < 0.0000000000000002 ***
sleep_hours 0.436876 0.077699 5.623 0.0000000617 ***
previous_scores 1.022061 0.008128 125.749 < 0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.995 on 203 degrees of freedom
Multiple R-squared: 0.9897, Adjusted R-squared: 0.9895
F-statistic: 6480 on 3 and 203 DF, p-value: < 0.00000000000000022Interpreting Coefficients
\[ \hat{Y} = b_0 + b_1X_1 + b_2X_2 + ... + b_kX_k \]
Focus on a specific predictor (e.g., \(X_1\))
For every 1 unit change in \(X_1\), there is a \(b_1\) unit change in \(Y\), holding all other predictors constant
Note: Properties of OLS still hold true
The sum of residuals will be 0
Each predictor (\(X\)) will be uncorrelated with the residuals
Holding Constant??? Wut
Reporting the results
We fitted a linear model (estimated using OLS) to predict performance_index
with hours_studied, sleep_hours and previous_scores (formula: performance_index
~ hours_studied + sleep_hours + previous_scores). The model explains a
statistically significant and substantial proportion of variance (R2 = 0.99,
F(3, 203) = 6479.94, p < .001, adj. R2 = 0.99). The model's intercept,
corresponding to hours_studied = 0, sleep_hours = 0 and previous_scores = 0, is
at -33.16 (95% CI [-34.75, -31.57], t(203) = -41.17, p < .001). Within this
model:
- The effect of hours studied is statistically significant and positive (beta =
2.85, 95% CI [2.75, 2.96], t(203) = 54.36, p < .001; Std. beta = 0.39, 95% CI
[0.37, 0.40])
- The effect of sleep hours is statistically significant and positive (beta =
0.44, 95% CI [0.28, 0.59], t(203) = 5.62, p < .001; Std. beta = 0.04, 95% CI
[0.03, 0.05])
- The effect of previous scores is statistically significant and positive (beta
= 1.02, 95% CI [1.01, 1.04], t(203) = 125.75, p < .001; Std. beta = 0.90, 95%
CI [0.88, 0.91])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald t-distribution approximation.Creating the Model
There can be many different ways in which we create and compare models with multiple predictors
- Simultaneous: Enter all variables in the model at once
- Usually the most conservative and defensible approach (unless there is theory to support a hierarchical approach)
- Hierarchically: Building a sequence of models where a single variable is included/excluded at each step
- This is hierarchical/stepwise regression. Different from HLM (Hierarchical Linear Modeling)
Model Selection (LSR 15.10)
How can we tell if one model is “better” than the other (it explains more variance in outcome)?
- Each predictor (or set of predictors) is investigated as to what it adds to the model when it is entered
- The order of the variables depends on an a priori hypothesis
The concept is to ask how much variance is unexplained by our model. The leftovers can be compared to an alternate model to see if the new variable adds anything or if we should focus on parsimony
Model Diagnostics
Checking the model (LSR 15.9)
Whenever we are looking at the regression model diagnostics, we are often considering the residual values. The types of residuals we can look at are:
Ordinary Residuals - Raw
Standardized Residuals
Studentized Residuals - Takes into account what the SD would have been with the removal of the \(i\)th observation
Model Checks - Outlier
We tend to look for various things that may impact our results through the lens of residuals
1) Outliers - variables with high Studentized Residuals
Model Checks - Leverage
We tend to look for various things that may impact our results through the lens of residuals
2) Leverage - variable is different in all ways from others (not just residuals)
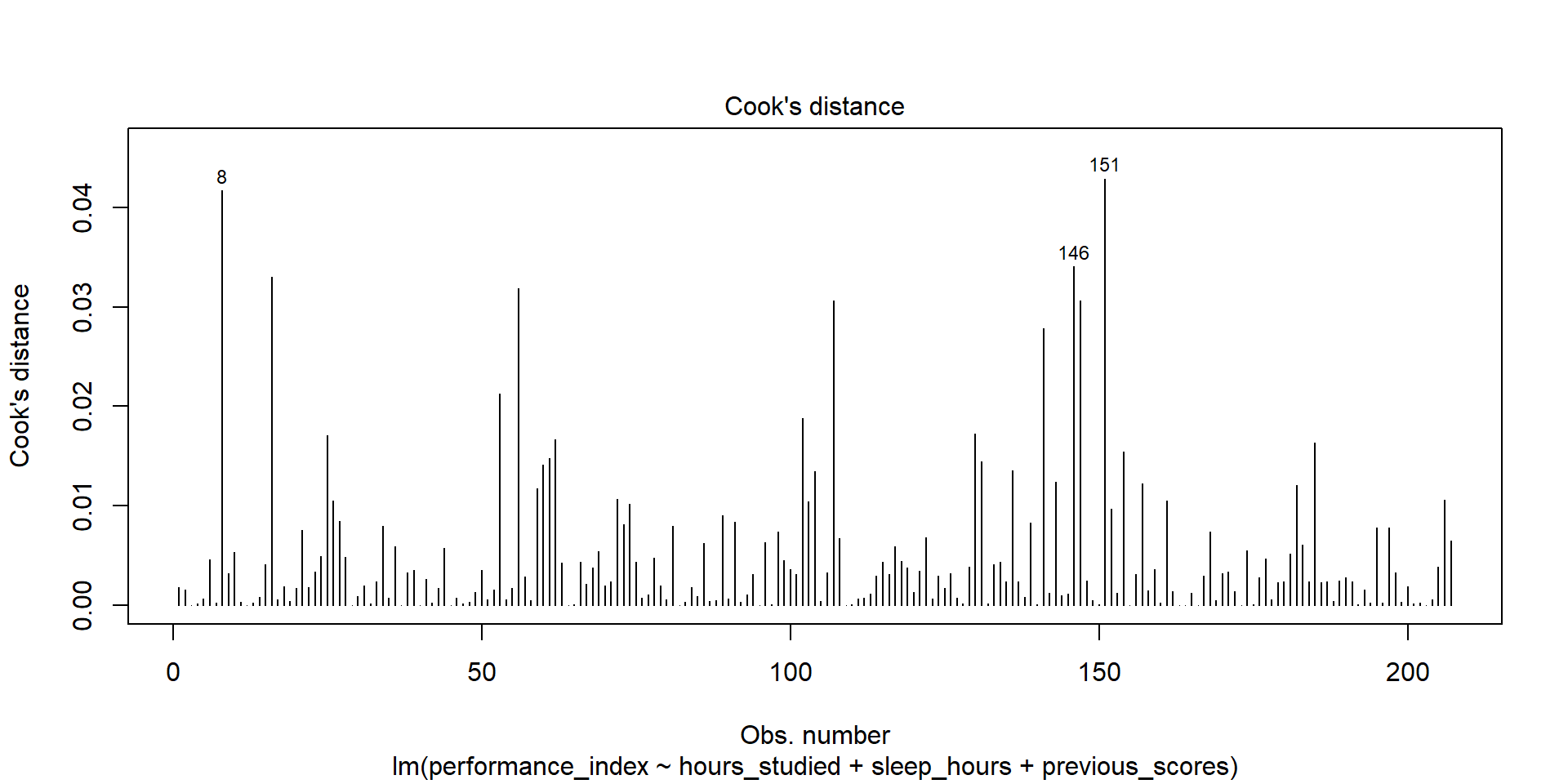
Model Checks - Influence
We tend to look for various things that may impact our results through the lens of residuals
3) Influence - outlier with high leverage (Cook’s Distance)
Model Checks - Plots
Checking Collinearity
We need to check to see if our predictor variables are too highly correlated with each other
To do so we use variance inflation factors (VIF)
- There will be a VIF that is associated with each predictor in the model
- Interpreting VIF (link) - Starts at 1 and goes up
- 1 = Not Correlated
- 1-5 = Moderately Correlated
- 5+ = Highly Correlated
RMarkdown
RMarkdown
This is something that we have been using this whole time!
The script that we use allows us to embed both regular text and code all into a single document. This is what we use for the labs with various levels of success when it comes to knitting the document
We’ll be spending more time exploring all the cool things that RMarkdown can do
Some Useful Resources
Ideas for topics to Highlight
Making sure we can have things knit
Code Chunk Options (Cheatsheet)